Introduction
Given a problem, the usual approach to solving the problem is to build several predictive models using different machine learning algorithms. When building the predictive models, we carefully choose the parameters of the models using the validation set and then evaluate and compare their performance on the test set to determine the best model. This is due to the fact that there is no single algorithm that will always give us the best model for every problem. Building an optimal predictive model with finite data is a challenging task. Machine learning algorithms come with different sets of assumptions that induce certain models. If the assumption about the data is wrong, it leads to underfitting and high bias error.
One can fine-tune the parameters to get a model with the highest validation accuracy. But finding the optimal parameters is an arduous task. In many cases, even after considerable fine-tuning, the model is still not good enough and fails on certain instances. Furthermore, in some applications such as medical and healthcare, datasets are not easy to obtain and the number of instances is typically limited. This further exacerbates the problem and the task of developing a reliable predictive model becomes somewhat infeasible. Evidently, a single model is not sufficient and there is a need for algorithms that can combine multiple models to produce better predictive models. Ensemble learning is an approach to combining the decisions of multiple models when predicting new instances [1]. The idea is that, if a model incorrectly predicts an instance, there may be another model that is able to get it right. Essentially, we want to leverage the strength of multiple models with different characteristics such that the models induce different outcomes so that they can complement each other.
Ensemble Diversity
An ensemble model supposes to perform better than any single model, that is, it should have a lower classification or regression error. We might think that a good ensemble model can be simply obtained by combining several accurate individual models. However, model accuracy is not the only consideration when building an ensemble model. More importantly, when we are building an ensemble model, the individual models or known as base models are diverse in the sense that they are making different predictions [2]. Otherwise, there would be no performance improvement even if the ensemble model is a combination of a large number of accurate models.
Building ensemble models is not an easy task. One simple approach to ensemble learning is to train multiple base models and assign a weightage to each model to indicate its importance. But this approach normally does not work because the models are trained for the same objective. The task becomes even more challenging when the same training set is used to train the models which could make them highly correlated. Furthermore, at the same time, we need to ensure the performance of the models must not very poor. Otherwise, the ensemble model’s performance would not improve or worsen.
Ensemble diversity refers to the differences among the individual models that make up the ensemble model. Several approaches have been used to achieve the goal of generating diverse models. The first approach is to use different learning algorithms to train the base models. Different learning algorithms have different inductive biases. If a base model fails to predict certain instances, other models might get them right due to their different assumptions about the data, which improves the ensemble performance. For example, one base model may be parametric (e.g. logistic regression) and others are non-parametric models (e.g. decision tree and non-linear support vector machine).
We can also build diverse base models using the same learning algorithm but with different parameters. For example, we can build different base models by varying the kernel functions in support vector machine, the number of nearest neighbors in k-nearest neighbors the tree depth and the minimum number of instances in a leaf node in decision tree [3]. If the models are trained using optimization techniques such as gradient descent in linear regression and neural network (this algorithm is discussed in a later chapter), the initial weights, learning rate and batch size are hyperparameters that can be varied to build diverse models [4].
Another approach to building diverse base models is sampling manipulation whereby the base models are trained using different training subsets. The subsets can be created using bootstrapping [5], a sampling method with replacement whereby instances are randomly drawn from the training set. Base models that are trained with different subsets are usually diverse. This ensemble learning is known as bagging which will be discussed later. The base models can also be trained sequentially whereby for each iteration (training), instances that are incorrectly predicted are given more emphasis in training subsequent base models. This ensemble learning is known as boosting.
Combination Methods
Given a set of base models, there are numerous methods to combine the predictions of the models into a single prediction. The commonly used methods are voting, linear combination and stacking.
Voting is used to combine base models that output class labels, thus it is applicable for classification ensembles. Voting combines the classification of base models by majority voting. Each base model outputs a particular class label and the class label with the most votes is chosen as the ensemble output. The voting ensemble is defined as:
![]()
where ![]() is the set of base models.
is the set of base models.
Linear combination is used to combine real-valued outputs such as class probabilities, thus it is applicable to both classification and regression ensembles. In linear combination method, each base model is assigned a weightage which implies the importance of the model. The combined probability of class ![]() of the models is defined as:
of the models is defined as:
![]()
where ![]() is the weightage of
is the weightage of ![]() -th base model and
-th base model and ![]() and
and ![]() . If the weights are equal
. If the weights are equal ![]() , this is a simple averaging. However, in practice, the base models would have different performances, hence the base models would have different weights, and finding the optimal weights manually is difficult if not infeasible [6]. One possible way to solve for
, this is a simple averaging. However, in practice, the base models would have different performances, hence the base models would have different weights, and finding the optimal weights manually is difficult if not infeasible [6]. One possible way to solve for ![]() is to assess the accuracies of the base models on the validation set.
is to assess the accuracies of the base models on the validation set.
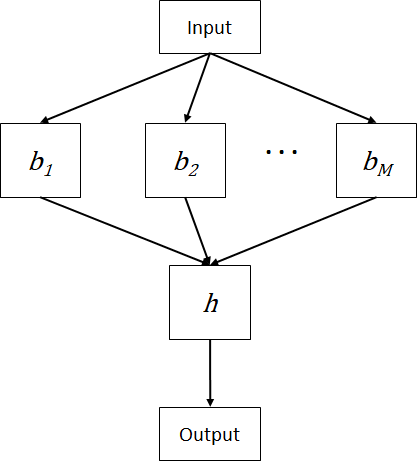
Stacking or staked generalization is a method that involves the use of machine learning algorithm to train a predictive model to combine the predictions of the base models [7]. The predictive model is referred to as the meta model. Specifically, the predictions of the base models serve as inputs to the meta model which is trained to optimally combine the predictions. In other words, the meta model learns the errors and corrects the biases of the base models. Figure 1 shows the stacking combination method whereby a meta model ![]() accepts the predictions of
accepts the predictions of ![]() base models to produce the prediction. The meta model can be built using any machine learning algorithm. But in practice, logistic regression and linear regression are often used as the meta model for classification and regression problems respectively.
base models to produce the prediction. The meta model can be built using any machine learning algorithm. But in practice, logistic regression and linear regression are often used as the meta model for classification and regression problems respectively.
The training is done in two phases. First, the training set is divided into two subsets, one is for training the base models and the other one is used to train the meta model. Using the first subset, the base models are trained and the base models should be as different as possible so that they will make different errors. The second training phase uses the second subset to train the meta model. The second subset is fed to the base model to produce a set of predictions which will be used as the training input for the meta model. Then, the ensemble stacking model is evaluated on the test set by feeding the data to the base models, collecting the predictions of the base models and feeding the predictions to the meta model to get the final predictions.
References
[1] D. Opitz and R. Maclin, “Popular Ensemble Methods: An Empirical Study,” Journal of Artificial Intelligence Research, vol. 11, pp. 169–198, Aug. 1999, doi: 10.1613/jair.614.
[2] P. Sollich and A. Krogh, “Learning with Ensembles: How over-Fitting Can Be Useful,” in Proceedings of the 8th International Conference on Neural Information Processing Systems, Cambridge, MA, USA, 1995, pp. 190–196.
[3] F. T. Liu, K. M. Ting, and W. Fan, “Maximizing Tree Diversity by Building Complete-Random Decision Trees,” in Advances in Knowledge Discovery and Data Mining, Berlin, Heidelberg, 2005, pp. 605–610. doi: 10.1007/11430919_70.
[4] J. F. Kolen and J. B. Pollack, “Back propagation is sensitive to initial conditions,” in Proceedings of the 1990 conference on Advances in neural information processing systems 3, San Francisco, CA, USA, Oct. 1990, pp. 860–867.
[5] B. Efron and R. J. Tibshirani, An introduction to the bootstrap. CRC press, 1994.
[6] Z.-H. Zhou, Ensemble Methods: Foundations and Algorithms, 1st ed. Chapman & Hall/CRC, 2012.
[7] D. H. Wolpert, “Stacked generalization,” Neural Networks, vol. 5, no. 2, pp. 241–259, Jan. 1992, doi: 10.1016/S0893-6080(05)80023-1.